Jan24 Leslie's new method on estimating rates of 3' end pre-mRNA cleavage is now published in RNA! Congrats to all!
Jan24 Welcome to GSBS rotation student Joe Paquette, woh will be rotating with us through March! He will be working on investigating the dynamics of small RNAs upon immune stimulation.
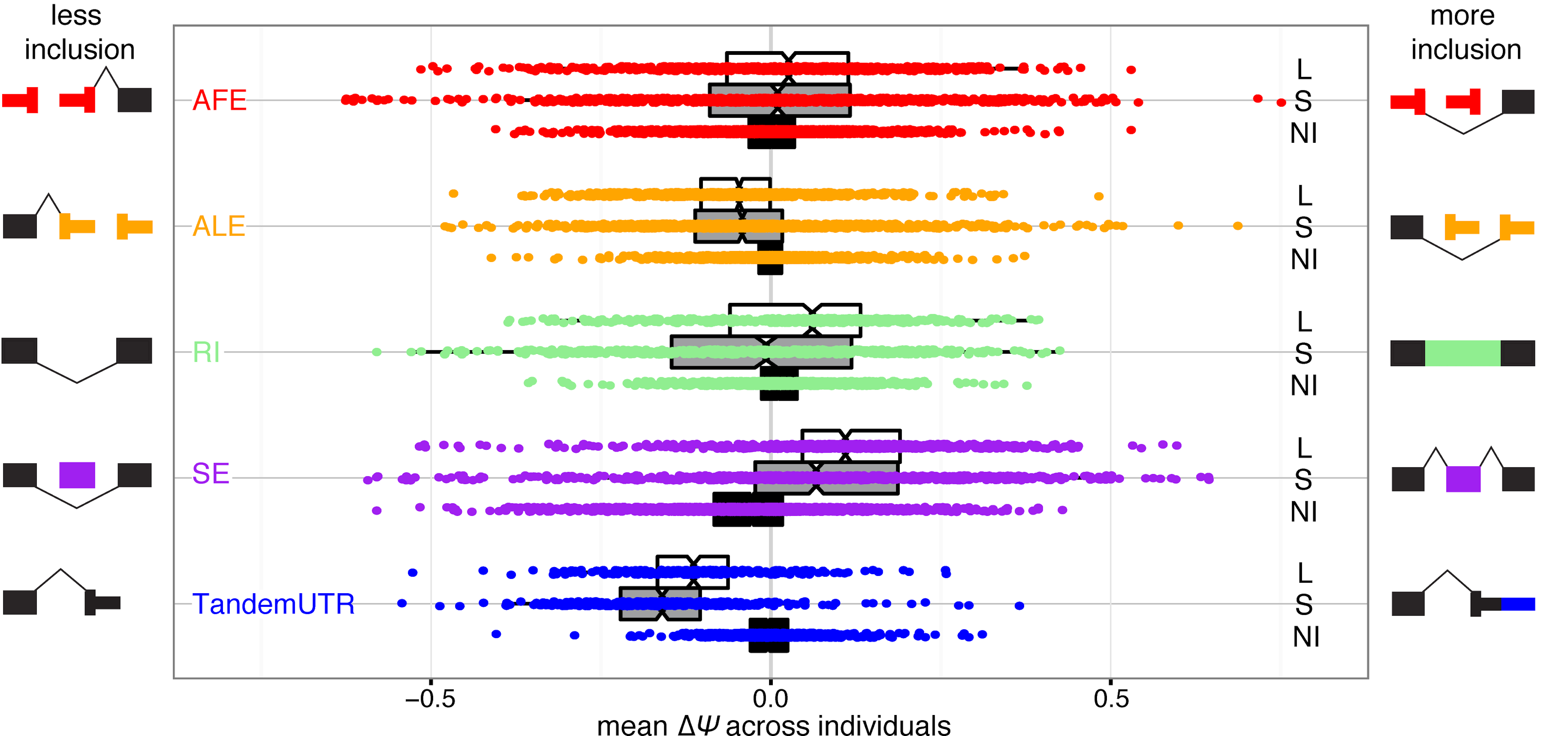
Jan24 PREPRINT ALERT! A long labor of love from Ezequiel and Christine and a result of our long-standing collaboration with the Fiszbein lab at BU. We describe our discovery of widespread positional coordination in the usage of mRNA start and end sites. We call this phenomenon the Positional Initiation Termination Axis (PITA) and characterize the functional impacts + potential molecular causes in our new study. Thanks to all those involved and especially to Sergey & Job from the Dekker lab for their help in shedding some mechanistic insight!
Nov23 Congrats to Valeria Sanabria for winning the Zelda Haidak Scholarship in Cell Biology, awarded to a UMass Chan graduate student aiming to establish a research career in cell biology. Read more about the news here!
Nov23 We welcome Valeria Sanabria as a new graduate student in the lab! She will be developing projects focused on leveraging long read sequencing to understand cryptic splicing.
Oct23 Welcome to GSBS rotation students Christable Darko and Natalie Haas, who will be rotating with us through October! They will be working on long-read sequencing studies and cryptic splicing analyses, respectively!
Sep23 Congrats to Jesse Lehman, who was awarded a spot on the competitive UMass Chan Innate Immunity Training Program T32 grant!
Aug23 The lab has been a new R01 from NGHRI, which is the chapter in our long-standing collaboration with the Engelhardt lab at the Gladstone Institute/Stanford! We will work on developing a statistical framework to understand how gene regulatory kinetics underlie alternative RNA isoform expression. RECRUITING POSTDOCS TO JOIN THIS COLLABORATION!
July23 PREPRINT ALERT! Congrats to Ezequiel and Rachel, who toiled away at sysmetically assess the accuracy of identifying mRNA terminal ends from long read sequencing data - lots of insights, advice, and cautious tails (pun intended).
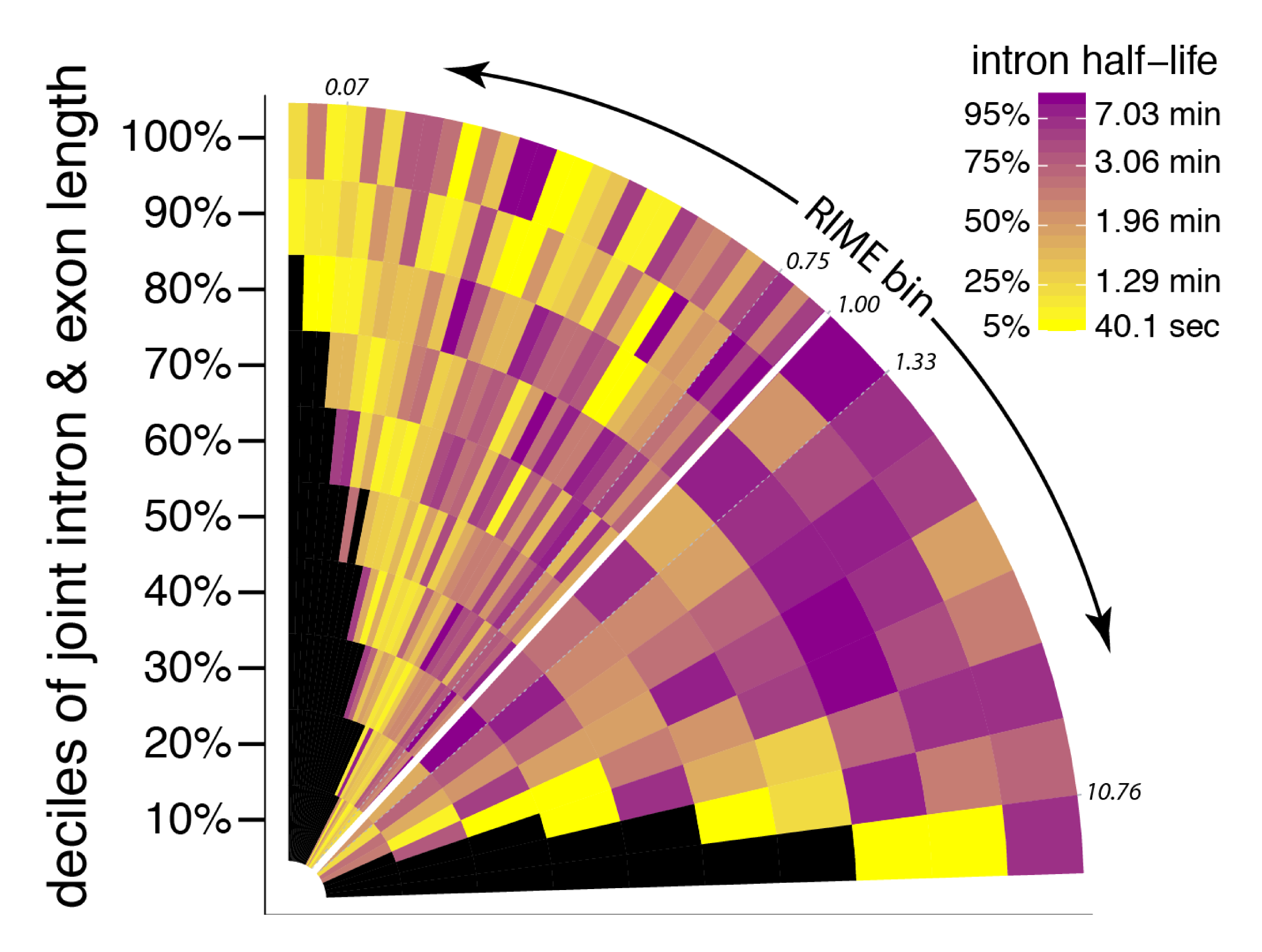
July23 PREPRINT ALERT! Congrats to Leslie, who completed a manuscript describing our new method to estimates rates of 3' end pre-mRNA cleavage - for the first time - and how/why cleavage rates vary in Drosophila S2 cells! Thanks to Ezequiel also for his contributions!
Jun23 Welcome back for Arvo Justice, our summer intern who just graduated high school and will spend the summer in our lab before leaving to start college at Johns Hopkins University!
031423 The Pai Lab Pi Day Pie Party has made a return - we were happy to take over the RTI monthly departmental tea time to share pie, pi jokes, and piRNA thoughts!
Mar23 Congrats to Jesse for passing his qualifying exam! He's back to lab with lots of excitement to pursue his thesis work on investigating the rates of transcription and splicing upon immune stimulation!
Mar23 The lab has been awarded a CAREER grant from the National Science Foundation! We're excited to use this funding to pursue our work on investigating the causes and consequences of cryptic splicing in mammalian cells!
Jan23 Lots of exciting collaborative papers have been published as the new year! Thanks to the Dekker (Valton et al. NSMB), Watts (Wang et al. NAR), and Fitzgerald (Vierbuchen et al. PNAS) labs on including us in their work and congrats to all the authors!
Jun22 We're excited to welcome Arvo Justice, a rising senior at Newton North High School, to the lab as a summer intern. He'll be working with Jesse on creating a lab resource for RNA-seq spikeins!
Mar22 Hannah and Yahui's study on circular isoforms of ANRIL, in collaboration with the Alonso lab at Weill Cornell, is now published in Scientific Reports! Congrats to all!
Mar22 We welcome a new graduate student, Jesse Lehman, who officially joined the lab after his recent rotation! He'll continue working on investigating RNA kinetics in the immune response, among other things we manage to excite him about!
31422 After a pandemic-induced break, we celebrated our third annual Pai Lab Pi Day Pie Party! Everyone brought in tasty pi pie goodies, with lots of creativity! Just within the lab for now, but hope to be back in a bigger way next year! #paipipie
Mar22 We bid farewell to Mustafa, who is leaving to pursue new adventures as a Research Scientist in the Weiss group at MIT.
Feb22 Welcome to GSBS rotation students Jesse Lehman and Vista Sohrab, who will be rotating with us through March! They will be working on investigating transcriptional kinetics in the immune response and the effects of U1 perturbation upon splicing kinetics, respectively.
Jan22 Welcome to Rachel Daniels, a senior researcher who is joining us from Harvard Chan School of Public Health. Rachel will be joint between the Pai and Karlsson labs - strengthening connections with BIB - and was recently awarded an R21 to study human hookworm genomics!
Jan22 Congrats to Hannah and Yahui for their preprint posted on bioRxiv, a great collaboration with the Alonso lab (formerly UMMS, now Weill Cornell)! We systematically identified, quantified, and characterized circular isoforms of ANRIL, a lncRNA involved in cardiometabolic disorders. And thanks to Ezequiel for his contributions!
Nov21 Welcome to GSBS rotation student Brad Class, who will be working on generation and analysis of nascent long-read data for the next two months!
May21 The lab's first pre-print is posted on bioRxiv, a great collaboration with our friends from the Fiszbein (BU) and Burge (MIT) labs! We describe the HITindex, a new method to classify exon categories using short-read RNA-seq, and how we used it to characterize hybrid internal-terminal exons. Congrats to Ezequiel, Athma, and all other authors!
Apr21 Congrats to Ezequiel for passing his qualifying exam! He's excited to continue work on the co-regulation of transcription and termination for his thesis research!
Jan21 Happy New Year! Here's hoping for a less crazy 2021! To help us achieve that, we welcome a new administrative coordinator, Kathy Sloan!
Nov20 Welcome to Mustafa Malik Ghulam, a postdoc joining us after completing his PhD at University of Sherbrooke in Quebec, Canada. Mustafa will be joint between the Pai and Sontheimer labs - strengthening connections with our next door neighbors!
Aug20 Congrats to Hannah and Leslie for passing their qualifying exams! They're excited to continue work on RNA processing kinetics - with Hannah focusing on mutation-altered kinetics in cancer cells and Leslie on how RNA processing might underlie the timing of the circadian clock for their respective thesis research.
May20 Welcome to MD/PhD rotation student Ayush Kumar, who will be working remotely with Eraj on noisy splicing analysis for the next month!
Apr20 Some good news among the whirlwind of 2020 - Ezequiel Calvo Roitberg has officially joined the lab for his thesis research! Welcome Ezequiel!
Mar20 Wet lab work has been put on pause due to the ongoing pandemic, but our computational work continues from home. Sadly the third annual Pai Lab Pi Day Pie Party has to be postponed - fingers crossed, we'll be back next year with a pie contest! #paipipie
Mar20 We've been funded by SLC6A1 Connect for a collaborative project with the Watts Lab to develop ASOs that target noisy splicing and up-regulate gene expression!
Feb20 We bid farewell to Paul Yan, who was instrumental to the lab's early days. We wish him well at his next adventure in industry!
Feb20 Welcome to GSBS rotation student Abigail Zeamer, who will be working on analysis and generation of nascent RNA-seq data for the next two months!
Dec19 Welcome to Eraj Khokhar, our first postdoc! Eraj comes to us after completing his PhD at The Jackson Laboratory in Maine.
Nov19 Welcome to GSBS rotation student Ezequiel Calvo, who will be working on optimizing our long-read nascent RNA-seq protocols over the next two months!
Sep19 We're excited to send Kevin off to graduate school! Luckily he's not going far - he just enrolled in the UMMS GSBS program, so we're looking forward to watching his graduate career bloom quite closely!
Jul19 The lab has our first Notice of Award! We have been awarded an R35 (MIRA) from NIH NIGMS to continue our work on investigating the temporal progression of RNA biogenesis and maturation!
Aug19 We were thrilled to contribute to a study developing Divalent-siRNA therapies for substantial huntingtin> knockdown as a therapeutic for Huntington's Disease, led by the Khvorova lab and now published in Nature Biotechnology.
Jul19 Congrats to Paul for passing his qualifying exam! He's excited to continue working on splicing kinetics over neuronal differentiation to understand molecular mechanisms underlying Huntington's Disease.
May19 Busy times in the Pai Lab this summer - Katy Monopoli (UMMS-WPI) will be joining us for a summer rotation and undergrads Parker Simpson (WPI) and Vista Sohrab (UMass Amherst) will be doing summer internships. Welcome!
Apr19 Hannah Macmillan and Leslie Torres Ulloa have officially joined the lab for their thesis research! We are excited to welcome them to lab!!
31419 Second annual Pai Lab Pi Day Pie Party was thrown in conjunction with the Garber Lab during the 3/18 RTI Teatime. Thanks to our many colleagues for showing up to enjoy pi[e] with us! #paipipie
Feb19 Welcome to rotation students Han Zhang (GSBS), Hannah Macmillan (GSBS), Jocelyn Pettito (WPI-UMMS), and Shaimae Elhajjajy (UMMS-WPI)! We will have a full house through March!
Dec18 Welcome to GSBS rotation students Leslie Torres Ulloa and Alex Park, both of whom will be working on computational rotations with us through January!
Sep18 Welcome to GSBS rotation student Wenjia Huang, who will be working with Paul for the month.
Sep18 New review on the dynamics of RNA processing upon perturbation of cellular conditions now published in WIREs RNA!
Aug18 Our paper on recursive splicing has been published in PLOS Genetics, with Paul and Athma as authors! Note the sister paper by Zhiping Weng's group in the same issue.
Jun18 Paul Yan joins the lab as our first graduate student! Paul has an undergraduate degree from UMass Lowell and is pursuing a Ph.D. in the UMass Med GSBS program.
Apr18 Kevin Fortier joins us as a research associate, previously having worked with Sean Ryder at UMMS! Kevin will be working on optimizing experimental protocols and ultimately automation of our basic workflow.
Apr18 Welcome to rotation students Eleni, Garhom, and Paul - looking forward to exploring exciting potential projects over the next couple months!
31418 First annual Pai Lab Pi Day Pie Party served as our lab warming. Thanks to our many colleagues for showing up to enjoy pi[e] with us! #paipipie
Jan18 Nida Javeed joins as our first lab member. Welcome Nida!
Jan18 The Pai Lab is officially open! Plenty of boxes to be unpacked and files to be moved!